Polygenic scores (PGS) are among the most contested offshoots that arose as a byproduct of genome wide association studies (GWAS), see for an example the tweet by Micheal Eisen included below. PGS are associated with lifting the effects off many or all genetic variants on a trait (usually a disease) from GWAS, in order to predict that trait in people that have yet to manifest it. But their original use in psychiatry was theoretical, brilliant and at least a decade ahead of its time. The lessons of that first paper where quickly buried by the hype, and in some cases real promise that came with predicting the onset of disease before it arises.


Michael Eisen isn’t entirely wrong obviously, there are plenty bad, misleading, and misguided papers based on polygenic scores. But, one promise I made myself when I set up this substack is that I wouldn’t waste my time and energy on bad science. Rather I want to highlight the (very) good stuff, the stuff that has you staring into the distance in awe at the busstop after reading, while the bus drives off without you.
The first paper I, and I think a lot of people in psychiatric genetics of a certain age, read on polygenic scores (PGS) was a paper by the International Schizophrenia Genetics consortium (open access version HERE).
ISC; Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 Aug 6;460(7256):748-52. doi: 10.1038/nature08185. Epub 2009 Jul 1. PMID: 19571811; PMCID: PMC3912837.Their GWAS, and their competitors GWAS of schizophrenia didn’t really yield a large number of genome wide significant SNPs, if any. A very disappointing results to some. Seven researchers within the consortium led an effort to see if the effect sizes found in early schizophrenia GWAS, would predict schizophrenia in a genotyped holdout sample. Their work was a direct test of a theorie first explicitly formulated in the 60’s the polygenic model of schizophrenia, schizophrenia s the end of a continous spectrum which in turn was the product of a large number of small genetic and environmental influences (Gottesman and Shield, 1967)
The researchers selected SNPs (single nucleotide polymorphism) binned according to their p-vlaue in the discovery GWAS and generate predictions of case/control status the holdout sample. This mean they made scores for SNPs with a p-value below 0.1 (pt < 0.10), below 0.2 (pt < 0.20), below 0.3 (pt < 0.30) etc etc. Their results where visualised in a manner that became synonymous with PGS research in psychiatric genetics for the next 5-8 years:
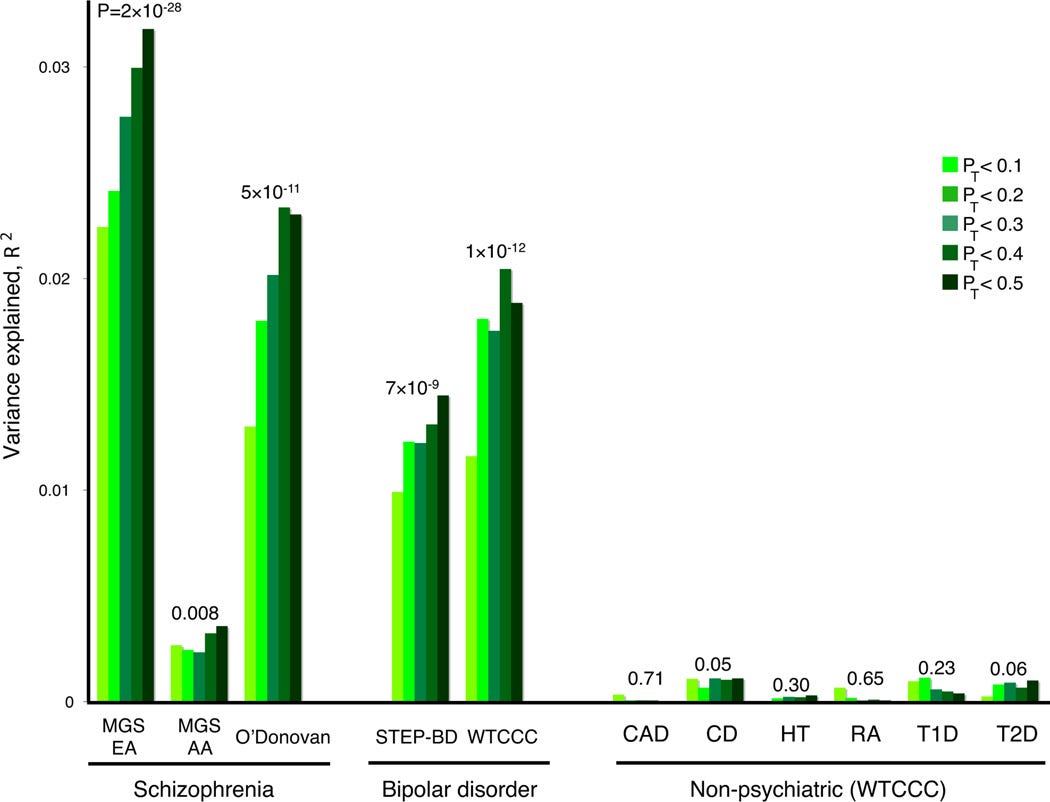
Based on the figure we can conclude 3 things: 1. the schizophrenia PGS is somewhat specific, because it didn’t predict non-psychiatric traits 2. it didn’t work as well in African Americans, a fact that could have alarmed researchers to the fact that their sampels weren’t diverse enough to generate health benefits for all, a decade ahead of the field. and 3. The scores also predicted bipolar disorder a genetic correlation that is among the highest in psychiatric genetics and calls into question some diagnostic practices, like the view that bipolar and schizophrenia are mutually exclusive or distinct entities. Finally the score based on a GWAS in males predicted schopghernia infemales very well, and vice versa.
But the authors did something far more sophisticated then just predict schizophrenia in an holdout sample, they simulated genetic architectures (a term that wouldn’t be popularised until way later) that could give rise to the observed nature of the polygenic score effects. They build a model in which they varied “Vm”, the heritability explained by all loci measure, or tggend by LD in the GWAS (they measured about 1 million common variants, most with a minor allele frequency > 5%), and varied the % of loci which has a causal effect form a hand full of loci, to 25%$ of the genome.
They then simulated how the PGS would have performed in each p-value bin given the grid models each with a specifi Vm and % of causal loci. For example if there had been 100 causal loci which explained ±30% of variance (Vm) in the risk for schizophrenia, these would almost all have to have had a p-value < 0.01 in the discovery GWAS and the score based on SNPp with p < 0.01 would have done really well in the holdout sample, adding more SNPs with less significant p-values would make the score perform worse (by adding noice). In their figure 3 they compare the empirically observed score performance across finely grained p-value inclusion criteria (left panel), to those the results expected given simulated models that are consistent with the empirical observation (middle panel) and results give models those that where not constent with the observed results (right panel)
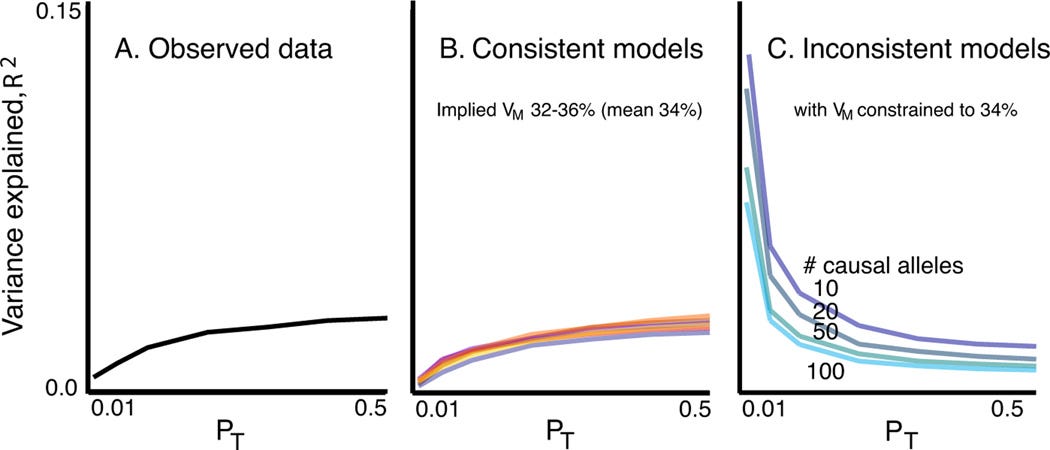
Their results where consistent withVm, now know as a SNP heritability (a term that didn’t exist yet) of 34% and between 6.25% and 25% of genome wide loci having some non-zero causal effect on schizophrenia (i.e a very very, almost imaginably, polygenic trait). The paper then proceeds to show how the causal alleles aren’t all going to be rare (< 5%) but rather are going to be spread over the entire allele frequency spectrum. The results preempted a decade of research, but to many (me included for a while) became synonymous with prediction.
You could imagine people setting out to estimate the same or similar parameters for all complex traits in the summer and fall of 2009, this could have accelerated several debates that played out in the field over the following decade.Their work reveals PGS have trouble generalising across ethnicities a problem if their ever going to be implemented in care, their results speaks to the polygenic on the nature of complex traits and so indirectly of what sample sizes we should expect our GWAS to need. However, PGS garnered their fame for something they didn’t do well (yet!) at all at the time, and for many traits still dosnt’t: prediction. Hwever even there reading the paper closely you learn that PGS will require incredibly large samples to work, and that both supporters and detractors should exert patience and withhold their final judgement on the utility of PGS, at least for another ±5-10 years and 3 orders of magnitude in sample size.
The paper is a true tour de force, had people realised the true potency of PGS for statistical genetics (as I am sure some did at the time!) their first decade may have ben less contentious. The paper effectively pre-empted some of the core notions studied in the super high profile papers listed below (which collectively could easily be the backbone of a syllabus of a grad level course on statistical genetics). Obviously these papers in turn had their own critical contributions to the field and may themselves feature in a future post.
Estimating the genetic correlation between bipolar disorder and schizophrenia was central in these widely read high impact papers:
Devlin, B., Kelsoe, J. R., Sklar, P., Daly, M. J., O'Donovan, M. C., Craddock, N., ... & Wray, N. R. (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature genetics, 45(9), 984-994.
Bulik-Sullivan, B., Finucane, H. K., Anttila, V., Gusev, A., Day, F. R., Loh, P. R., ... & Neale, B. M. (2015). An atlas of genetic correlations across human diseases and traits. Nature genetics, 47(11), 1236-1241.
Estimating Va, or SNP heritability is deemed so innovative that it led to these high impact papers:
Golan, D., Lander, E. S., & Rosset, S. (2014). Measuring missing heritability: inferring the contribution of common variants. Proceedings of the National Academy of Sciences, 111(49), E5272-E5281.
Bulik-Sullivan, B. K., Loh, P. R., Finucane, H. K., Ripke, S., Yang, J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, ... & Neale, B. M. (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics, 47(3), 291-295.
Estimating polygenicity (% of causal loci) has given rise to a decade of sophisticated statistical genetic technique :
Zhang, Y., Qi, G., Park, J. H., & Chatterjee, N. (2018). Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nature genetics, 50(9), 1318-1326.
Holland, D., Frei, O., Desikan, R., Fan, C. C., Shadrin, A. A., Smeland, O. B., ... & Dale, A. M. (2020). Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genetics, 16(5), e1008612.
O'Connor, L. J., Schoech, A. P., Hormozdiari, F., Gazal, S., Patterson, N., & Price, A. L. (2019). Extreme polygenicity of complex traits is explained by negative selection. The American Journal of Human Genetics, 105(3), 456-476.

